Main content
Top content

Ultraviolet light generation using infrared femtosecond laser pulses and nonlinear niobate nanoparticles (unpublished results)
Visualization and manipulation of microcompartments at ultrahigh resolution
Coordinator: Jacob Piehler
The focus of research field B is the advancement and adaptation of superresolution microscopy techniques in order to resolve microcompartment dynamics. The effective combination with complementary imaging modalities is another goal. To these ends these we will need
- access to state-of-the-art high resolution fluorescence microscopy techniques
- single molecule localization microscopy for super-resolving three-dimensional imaging and its adaptation to the investigation of microcompartments
- multi photon excitation microscopy techniques focusing upon varying non linear optic (NLO) nanomaterials
- electron microscopy for three-dimensional ultrastructural imaging, the localization of single protein complexes and correlative microscopy (CLEM)
Projects
- B1 - Mechanisms of development and degeneration in neuronal cells
- B2 - Synthesis of anorganic nano crystals
- B3 - Signal transduction in the yeast cell wall integrity pathway
- B4 - Pathogen-host interactions
- B5 - Dynamics and hemostasis of membran lipids
- B6 - Ultrafast physics - Visualization and manipulation by femtosecond-laser microscopy
- B7 - Molecular mechanisms of immune activation - S100A8/A9 proteins: Receptors and signal transduction
- B8 - Metabolic cross-feeding interactions among bacteria - Contact-dependent interactions among bacteria
- B9 - Extracellular matrix and organogenesis
- B10 - Extracellular matrix and organogenesis
- B11 - Signal propagation across biological membranes - Spatiotemporal organization of signaling complexes
- B12 - Bioorganic synthesis of nucleolipids as "antitumor small molecules"
- B13 - Dynamics and assembly of cytoskeleton associated complexes
- B14 - Molecular Basis of Behaviour - Interaction of stress and energy metabolism in clinically relevant animal models
- B15 - Membrane remodeling in the endolysosomal system
- B16 - Biomolecules at interfaces/Spatiotemporal organization of canonical Wnt signaling
- B17 - Plant Developmental Genetics - Transcriptional regulation in the nucleus
High and super-resolving microscopy techniques in CellNanOs
| Technology | Person in charge |
|---|---|
| Confocal microscopy including MPE, FLIM, FCS | Brandt, Hensel, Piehler |
| Spinning disk microscopy | Hensel |
| Deconvolution microscopy | Ungermann |
| Single molecule localization microscopy (PALM/STORM/TALM) | Piehler |
| Scanning force microscopy with fluorescence microscopy | Hensel, Piehler |
Multi-photon microscopy techniques | Imlau |
| Upconversion microscopy | Haase, Piehler |
| Transmission electron microscopy | Hensel, Paululat |
| Scanning electron microscopy | Paululat |
Brandt Lab, Biology
Mechanisms of development and degeneration in neuronal cells
Description coming soon.
Haase Lab, Chemistry
Synthesis of anorganic nano crystals
Description coming soon.
Heinisch Lab, Biology
Signal transduction in the yeast cell wall integrity pathway
Description coming soon.

Three-dimensional super-resolved reconstruction of the type III secretion system in Salmonella via dSTORM (rechts). Left: raw data as maximum intensity projection (Hensel lab, unpublished).
Hensel Lab, Biology
Pathogen-host interactions
Description coming soon.
Holthuis Lab, Biology
Dynamics and hemostasis of membran lipids
Description coming soon.
Imlau lab, Physics
Ultrafast physics - Visualization and manipulation by femtosecond-laser microscopy
The Imlau research group studies new methods for the optical control of nanophotonic probes in cellular microcompartments. Particular focus lies upon the localized generation of ultraviolet light (< 400 nm) using harmonic nanoparticles that allows for high-resolution (< 100 nm) photomanipulation in cells. We are interested in the efficiency of novel photo-physical processes, e.g. nonlinear optical resonances, within tailored harmonic nanoparticles and the use of femtosecond laser pulses. According to our expertise, the latter can be precisely controlled with respect to pulse shape, complex pulse trains, and wavelength.
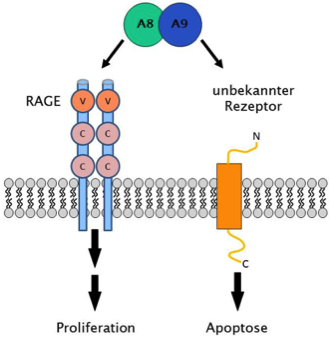
Via different receptors, which in part still need to be identified, S100A8/A9 can exert both proliferation and apoptosis in target cells (Ghavami et al., BBA 2008)
Kerkhoff lab, Human Sciences
Molecular mechanisms of immune activation - S100A8/A9 proteins: Receptors and signal transduction
The Kerkhoff group investigates the signal transduction of the calcium-binding complex S100A8/A9 after its binding to distinct receptors by which either proliferation or apoptosis are induced in target cells (cancer cells). We employ current molecular biological techniques and biochemical methods for the identification and analysis of protein-protein interactions, which will be explored with respect to spatiotemporal dynamics by advanced and superresolution imaging techniques. Thereby we intend to identify the corresponding receptors, to quantify protein-protein interactions at the plasma membrane, and to analyze the underlying signal transduction pathways.
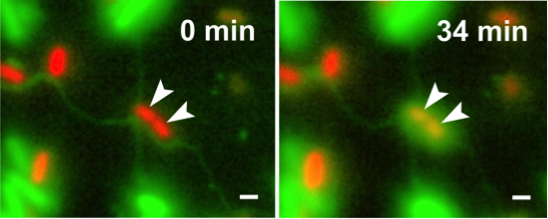
Escherichia coli (green cells) uses intercellular nanotubes to transfer cytoplasmic materials to Acinetobacter baylyi (red cells) (Pande et al., Nat Commun 2015)
Kost lab, Biology
Metabolic cross-feeding interactions among bacteria - Contact-dependent interactions among bacteria
The Kost group investigates contact-dependent metabolic interactions among bacterial cells. We study lipid-based nanotubes that auxotrophic bacteria form to derive amino acids from the cytoplasm of other bacterial cells. The project aims at investigation of the molecular and physiological underpinnings of intercellular nanotubes, particularly focusing upon functional aspects. To this end, nanotube-forming cells are imaged using high- and super-resolution microscopy and electron microscopy to shed light on the molecular machinery that underlies metabolite transport.
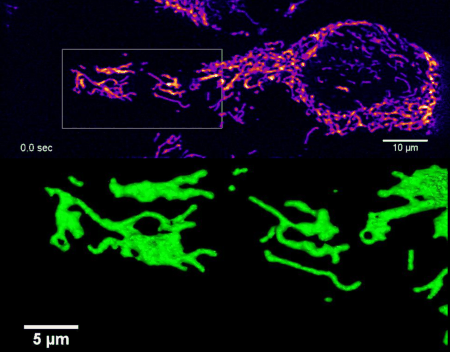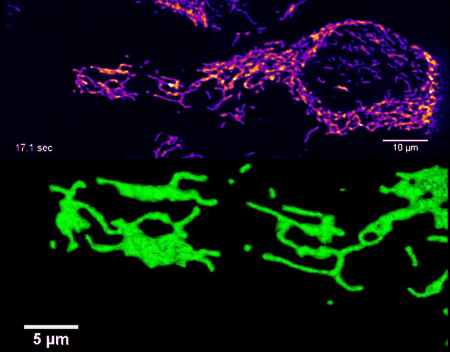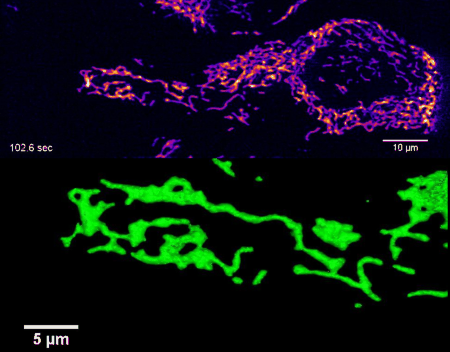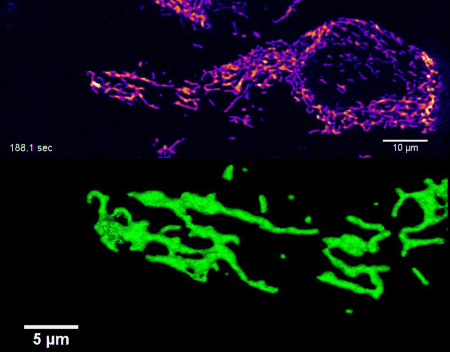
High-resolution four-dimensional imaging of the mitochondrial network in living HeLa cells by lattice light sheet microscopy (unpublished results)
Kurre lab, Biology
Superresolution fluorescence microscopy - Multimodal fluorescence microscopy at highest spatial and temporal resolution
We design and establish novel fluorescence microscopy techniques for live cell imaging in close collaboration with research groups from the departments of biology, chemistry and physics. To this end, we develop imaging protocols and workflows which are optimally tailored for different biological specimen and imaging systems. Our goal is to provide a broad user group with state-of-the-art multimodal microscopy systems for live cell imaging at highest spatial and temporal resolution. Our research focus is on multicolor single molecule localization microscopy as well as on the combination of structured illumination with (lattice) light-sheet microscopy.
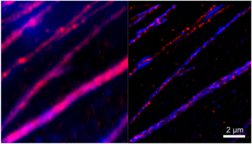
Co-organization of pericardin (red) and collagen fibers (blue) in the extracellular matrix of heart tissue in Drosophila visualized via dSTORM (right). Diffraction limited image (left) (Paululat lab, Piehler lab, unpublished).
Paululat Lab, Biology
Extracellular matrix and organogenesis
Description coming soon.
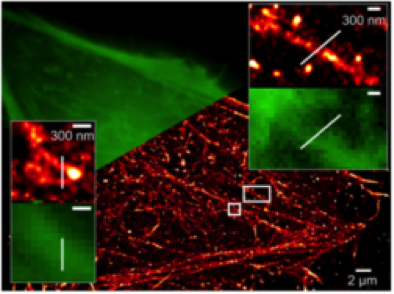
Super-resolved image of the cortical actin cytoskeleton in living cells via direct STORM (Wilmes et al., Angew. Chem. 2012)
Piehler lab, Biology
Signal propagation across biological membranes - Spatiotemporal organization of signaling complexes
The Piehler group studies the assembly and dynamics of cytokine receptor complexes in the plasma membrane of living cells by single molecule microscopy. Using posttranslational conjugation techniques, target proteins are labeled with photostable dyes or luminescent nanoparticles (cf. research field A). Their co-organization as well as interactions are then probed by single molecule localization. Thus, we aim to unravel the role of plasma membrane microcompartmentation in the spatiotemporal organization of signaling complexes and its role in regulating signaling specificity. Currently, lattice light sheet microscopy is being implemented to explore signal propagation within the cell.
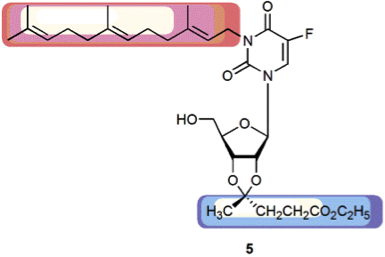
Novel nucleolipid with cytotoxic activity against several human tumor cell lines, but not against PMA-differentiated THP-1 macrophages (Knies et al., Chem Biodiv. 2015)
Rosemeyer lab, Chemistry
Bioorganic synthesis of nucleolipids as "antitumor small molecules"
The Rosemeyer group is heading at the combinatorial bioorganic synthesis of nucleolipid hybrid molecules, composed from either canonical nucleosides as well as nucleoside antimetabolites or natural or synthetic lipid molecules. The resulting compounds are investigated with respect to their cytostatic/cytotoxic activity a) towards human tumor cell lines of the NCI 60 panel, as well as b) against glioblastoma tumor cells. By comparison of conformational parameters of the substances – derived from detailed NMR analyses – with their anticancer activity, a quantitative structure-activity relationship (QSAR study) will be established.
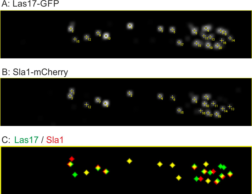
Co-localisation analysis via detection and tracking of endocytic events in Ashbya gossypii of the regulators Las17 and Sla1.
Schmitz Lab, Biology
Dynamics and assembly of cytoskeleton associated complexes
The Schmitz lab is working on changes of the actin cytoskeleton during polar growth and morphogenesis. Fast and directed growth modes are studied in fungal modelsystems. These studies involve processes of early endocytosis and the metamorphosis of the spindle pole body from a microtubule- to an actin organization center during sporulation. Both dynamic processes are studied primarily by light-microscopy in living cells. Precise evaluation of these dynamic processes will be performed by specifically adapted software that has been developed by the CALMOS. Additionally optogenetic methods will be tested and developed in this project.
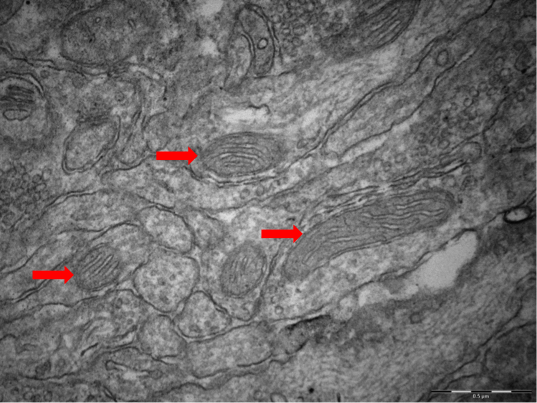
Electron microscopy image of mitochondria in the hippocampus of a "High Reactivity mouse"
Touma lab, Biology
Molecular Basis of Behaviour - Interaction of stress and energy metabolism in clinically relevant animal models
Research of the Touma group addresses the interactions of stress with alterations in neuronal and peripheral energy metabolism. What role do they play in the aetiology of diseases? Mechanistic links between a dysregulated stress hormone system and variations in metabolic functions (changed functionality of microcompartments and important cell organelles such as mitochondria) have been shown in patients and animal models of psychiatric disorders, ultimately resulting in accelerated cell ageing. We use the stress reactivity mouse model to unravel the underlying risk factors and pathomechanisms. Imaging with highest spatial resolution and advanced data processing techniques will allow us to elucidate the cellular determinants of stress regulation in our animal models.
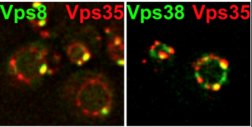
Relative distribution of endosomal proteins. The indicated proteins were genomically tagged with GFP and mCherry tags and monitored by fluorescence microscopy. The ring like structures are yeast vacuoles (Arlt et al., Mol Biol Cell, 2015).
Ungermann Lab, Biology
Membrane remodeling in the endolysosomal system
The Ungermann lab is interested in the fusion of membranes along the endolysosomal pathway in yeast and higher eukaryots. The group has developed protocols to visualize microcompartments of the vacuole, an to follow and manipulate the endocytic transport. High resolution of structures on both compartments are investigated by electron microscopy. The group develops novel techniques to monitor the spatio-temporal changes of endosomal and vacuolar structures in collaboration with other groups of the CellNanOs.
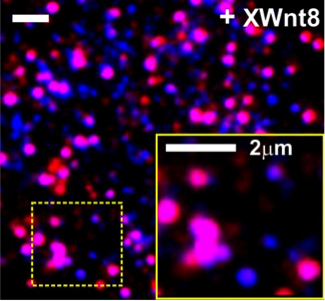
Single molecule fluorescence imaging reveals Wnt-induced formation of signalosomes in the plasma membrane of living HeLa cells (unpublished results)
You lab, Biology
Biomolecules at interfaces/Spatiotemporal organization of canonical Wnt signaling
The You group aims at exploration of the spatiotemporal regulation of canonical Wnt signaling. This is a key regulatory hub in stem cell differentiation and embryonic development. The molecular interplay of the numerous molecular players involved in the formation of the so-called ‘Wnt-signalosome’ remains largely elusive. By using advanced multicolor single molecule fluorescence imaging as well as protein micropatterning in the plasma membrane of living cells, we will dissect the molecular and cellular determinants governing assembly, dynamics and signaling of the Wnt signalosome.
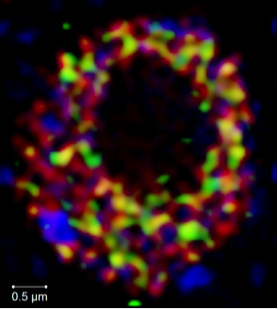
Resolving the subnuclear localization of ROXY1 (green), RNA polymerase II (red) and heterochromatin (blue) by structured illumination microscopy (SIM) (unpublished results)
Zachgo lab, Biology
Plant Developmental Genetics - Transcriptional regulation in the nucleus
The Zachgo lab investigates the function of ROXYs, land plant-specific glutaredoxins (GRX), which regulate plant developmental processes. ROXYs interact in the nucleus with the bZIP TGA transcription factors, which therefore represent potential targets for posttranslational modifications. Nuclear protein-protein interactions are analyzed by high- and superresolution fluorescence microscopy of fixed and live cells. Lattice light-sheet microscopy will be used to resolve the spatial organization of these proteins within the nucleus to gain insight into the dynamics of transcriptional interaction networks in nuclear microcompartments.

