Main content
Top content
Image processing, modeling and simulation
Coordinator: Stefan Kunis
Research field D is devoted to the development of algorithms and methods within the framework of biological problems in close collaboration of mathematicians, physicists and cognitive scientists. Aims include the
- quantitative evaluation and visualization of data from advanced imaging techniques
- identification of causality and correlations in complex data sets
- mechanistic description of molecular processes up to complete microcompartments.
Projects
- D1 - Mechanisms of development and degeneration in neuronal cells
- D2 - Ultrafast nonlinear optical spectroscopy
- D3 - Neurophysiological basis of cognitive functions - Visual processing and embodied cognition
- D4 - Analysis, algorithms, signal and image processing
- D5 - Image processing and visualization of complex multidimensional data
- D6 - Statistical physics
- D7 - Bioinformatics
- D8 - Signal propagation across biological membranes - Surface architectures for quantitative interaction analysis
- D9 - Statistical model and hypothesis formulation
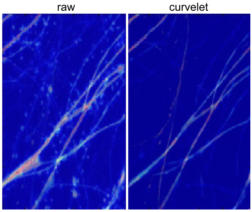
Neuronal morphology of brain sections of transgenic mice. Left: raw data, right: dendritic spines after curvelet analysis (Brandt lab, Kutyniok lab, unpublished).
Brandt Lab, Biology
Mechanisms of development and degeneration in neuronal cells
Description coming soon.
Imlau Lab, Physics
Ultrafast nonlinear optical spectroscopy
Description coming soon.
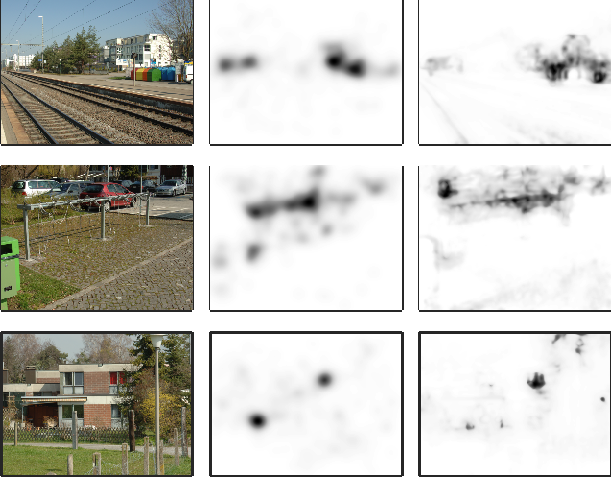
Comparison of visual exploration of natural stimuli (right) with model predictions (center) for natural stimuli (left) (Wilming et al., PLoS One 2011).
König lab, Cognitive Science
Neurophysiological basis of cognitive functions - Visual processing and embodied cognition
The research group of Peter König investigates multisensory integration and sensorimotor coupling with a variety of experimental and theoretical methods. This includes on one hand the quantitative analysis of subjective perception, eye tracking, EEG, MEG and fMRI. Furthermore, modeling of neuronal systems in the contest of embodied perception and processing of natural stimuli by neuronal networks and hierarchical Bayes models. In close interaction of experimental and theoretical work hypotheses are tested and successively refined. Results of this work are, as far as possible, translated to real world applications and have initiated founding of two spin-off companies.
Kunis Lab, Mathematics
Analysis, algorithms, signal and image processing
Description coming soon.
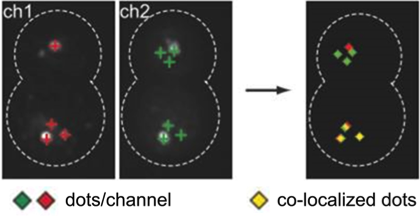
Automated single cell co-localization analysis (ImageJ plugin) to identify spatiotemporal organization of endosomal markers in yeast cells (Arlt et al., Mol Biol Cell, 2015).
Kurre lab, Biology
Image processing and visualization of complex multidimensional data
Complex multidimensional image data sets need a well-balanced post-processing strategy before an appropriate 3D visualization is feasible. We provide diverse tools for image deconvolution as well as volume and surface rendering in order to visualize dynamic processes at highest contrast. Furthermore, we develop tailored image processing tools for diverse scientific problems (protein co-localization, quantification of morphological changes, feature segmentation, etc.). Our aim is to integrate these tools into well-established software such as ImageJ or Matlab to increase usability and to extend user group.
Maaß Lab, Physics
Statistical physics
Description coming soon.
Mulkidjanian Lab, Physics
Bioinformatics
Description coming soon.
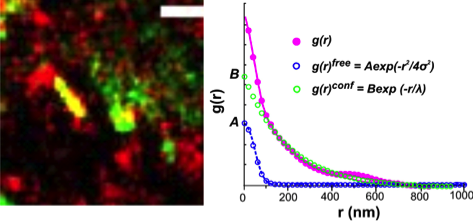
Pair-correlation tracking and localization microscopy (pcTALM) analysis of effector recruitment to a quantum-dot labeled cytokine receptor signaling complex (You et al., Anal Chem 2014)
Piehler lab, Biology
Signal propagation across biological membranes - Surface architectures for quantitative interaction analysis
For the analysis of images obtained by single molecule localization microscopy, the Piehler group has developed an integrated evaluation software. Next to the rendering and analyzing single molecule localization-based superresolution images, the characterization of diffusion dynamics and interactions by spatial and spatiotemporal correlation techniques is a particular focus of this work. In collaboration with the Kunis group, these efforts are pushed towards implementing fast 3D superresolution imaging in living cells by single molecule localization and structure illumination.
Pipa Lab, Cognitive Science
Statistical model and hypothesis formulation
Description coming soon.

