Main content
Top content
Qualitative and quantitative analytics of composition and interactions
Coordinator: Heinz-Jürgen Steinhoff
Within research field C we will develop and share techniques to dissect the protein and lipid compositions of microcompartments as well as identify and quantify molecular interactions. Specific objectives include
- the development and adaptation of mass spectrometric analyses of proteins and lipids in combination with optical and magnetic methods in order to selectively manipulate and isolate subcellular samples
- the extension of quantitative interaction analytics with purified protein as well as native cellular samples
- the development of quantitative binding assays for the localization and quantification of interactions in living cells
- the application and further development of structural biology techniques up to the investigation of protein conformations and conformational dynamics in their native environment.
Projects
- C1 - Molecular Membrane Biology - Mechanisms of sphingolipid and sterol homeostasis in Sacchomyces cerevisiae
- C2 - Signal transduction in the yeast cell wall integrity pathway
- C3 - Pathogen-host interactions
- C4 - Metabolic cross-feeding interactions
- C5 - Structural Biology of Molecular Machines - Regulation of signal and transport processes
- C6 - Electronic Transport - Ultra-Sensitive Sensors for Molecular and Nanoscale Phenomena
- C7 - Signal propagation across biological membranes - Surface architectures for quantitative interaction analysis
- C8 - Mesoscopic structure formation - Functional micro- and nanostructured surfaces
- C9 - Macromolecular structure - EPR spectroscopy of membrane proteins
- C10 - Membrane remodeling in the endolysosomal system
- C11 - Surface biofunctionalization for quantification and manipulation of cell signaling
Qualitative and quantitative interaction analytics in the CellNanOs
| Technique | Person in charge |
|---|---|
| Mass spectrometry (MALDI/ESI) | Ungermann |
| Quantitative interaction analysis at surfaces | Piehler |
| Scanning force microscopy | Piehler, Heinisch |
Protein crystallization | Kümmel |
| Electron spin resonance spectroscopy | Steinhoff |
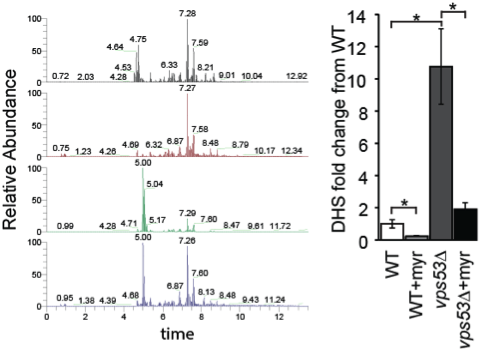
Analysis of sphingolipid levels from different yeast mutants by mass spectrometry based lipidomics (Fröhlich et al., Elife 2015)
Fröhlich lab, Biology
Molecular Membrane Biology - Mechanisms of sphingolipid and sterol homeostasis in Sacchomyces cerevisiae
Research of the Fröhlich group is focused upon sphingolipid and sterol homeostasis in the model organism Saccharomyces cerevisiae. To this end we investigate the sorting of lipids throughout the endocytic pathway and how these processes are regulated. In addition to the classic methods of cell biology we use proteomics, lipidomics, and systematic screenings. Unraveling these basic mechanisms will be the foundation to understand neurodegenerative disorders which are caused by the dysregulation of sphingolipid and sterol homeostasis.
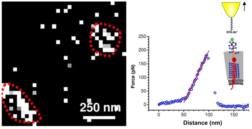
Cluster mapping of sensor complexes at the cell surface of yeast cells (left) and mechanical properties of single sensors measured via atomic force microscopy and spectroscopy (right) (Dupres et al., Nat. Chem. Biol., 2009).
Heinisch Lab, Biology
Signal transduction in the yeast cell wall integrity pathway
Description coming soon.
Hensel Lab, Biology
Pathogen-host interactions
Description coming soon.
Kost lab, Biology
Metabolic cross-feeding interactions
The Kost group studies cooperative cross-feeding interactions among different bacterial cells. For this, metabolite concentrations in- and outside bacterial cells as well as activities of the underlying metabolic pathways need to be determined. The group establishes protocols to accurately quantify metabolite concentrations using tools of analytical chemistry or genetically-encoded, intracellular biosensors. By analyzing both fluxes through the cells’ metabolic networks and the transcriptomes of individual cells or entire consortia, quantitative insights into the focal metabolic interactions can be derived.
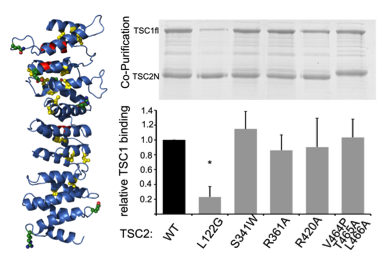
The structural analysis of the tumor suppressor TSC2 (left) reveals the mechanism of pathogenic mutations that cause tuberous sclerosis (right) (Zech et al., JBC 2016)
Kümmel Lab, Biology
Structural Biology of Molecular Machines - Regulation of signal and transport processes
The Emmy-Noether research group AG Kümmel is interested in the coordination, regulation and crosstalk between intracellular transport and signal transduction. We focus upon the structural investigation of proteins involved in these processes. The combination of X-ray crystallography with electron microscopy, biophysical and biochemical methods allows to visualize protein structures at atomic level and to explain their function. Furthermore, a detailed understanding of protein structure enables the investigation of the role of these proteins both under physiological and pathological conditions.
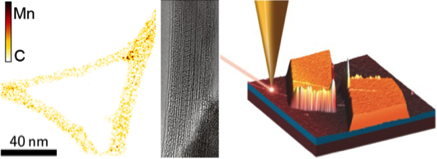
HRTEM images of {Mn4}-complexes on CNTs (left) and of C60 within CNTs (center). Right: AFM image of a CNT-FET with sketch of a TERS tip
Meyer lab, Physics
Electronic Transport - Ultra-Sensitive Sensors for Molecular and Nanoscale Phenomena
The group of Carola Meyer investigates upon electronic transport in carbon nanotubes (CNTs) functionalized with molecules. We focus on molecular interactions to gain a deeper understanding of spin interactions and molecular dynamics in magnetic and biomolecules. This includes the fabrication of nano-devices, the synthesis and chemical functionalization of the CNTs, and their characterization using cutting edge microscopy and spectroscopy. Electronic transport is investigated in quantum transport, noise and magnetoresistance measurements.
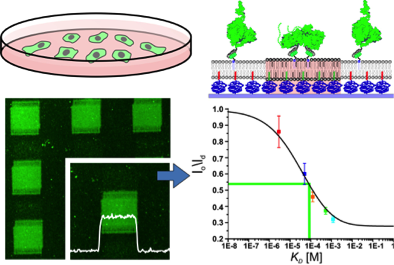
Quantification of low-affinity protein interactions by tethering to phase-separated plymer-supported membranes (Beutel et al., ACS Nano, 2015)
Piehler lab, Biology
Signal propagation across biological membranes - Surface architectures for quantitative interaction analysis
The Piehler group develops surface modification techniques for functional tethering of proteins including the reconstitution of integral membrane proteins into polymer-supported membranes. By combination of microstructured surface functionalization with optically or chemically switchable molecular recognition, spatiotemporally controlled organization of proteins on surfaces and in membranes has been achieved. Recently, this method has been extended to capturing proteins in the plasma membrane of living cells. These surface architectures offer versatile opportunities for quantitative biomolecular interaction analysis by a broad spectrum of spectroscopic and microscopic detection techniques as established in the group.

Scanning electron microscopy image showing a fibroblast grown on an artificial array of aligned polymer nanorods (Grimm et al., J. Mater. Chem. 2010)
Steinhart lab, Chemistry
Mesoscopic structure formation - Functional micro- and nanostructured surfaces
The Steinhart group investigates upon the design of topographically patterned surfaces to be used as synthetic biological interfaces. The topographic surface patterns may be combined with nanoscopic fine structures which represent additional functional hierarchical structure levels. Examples for surfaces with topographies tailored such as to control cellular interactions or adhesive properties include nanorod arrays and monolithic microsphere arrays. Examples for additional hierarchical structure levels with nanoscale dimensions are continuous pore systems enabling supply of liquid to synthetic biological interfaces and gold nanostructures for surface enhanced Raman spectroscopy.
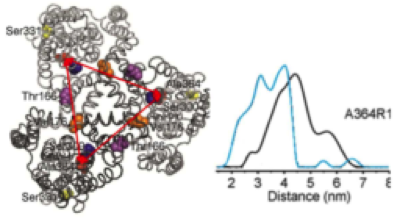
Distance distribution of the aspartate transporter Glt(Ph) in the presence (blue) and absence (black) of substrate as determined by DEER (Hänelt et al., Nat. Struct. Mol. Biol. 2013)
Steinhoff lab, Physics
Macromolecular structure - EPR spectroscopy of membrane proteins
The Steinhoff group studies the conformation and dynamics of biomolecules in order to understand their function on the atomic level. The group develops and applies advanced electron paramagnetic resonance (EPR) and double electron-electron resonance (DEER) methods in combination with site directed spin labeling, which are complemented by molecular dynamics simulations. The group focuses on exploring the functional mechanisms of transmembrane transport and signaling.
Ungermann Lab, Biology
Membrane remodeling in the endolysosomal system
The Ungermann lab is interested in the fusion of membranes of the endolysosomal pathway in yeast and higher eukaryots. The group had developed protocols to purify and characterize large protein complexes involved in membrane fusion and membrane remodeling at endosomes and vacuoles. The group has established new in vitro assays to analyze the membrane association and the subsequent membrane fusion, and has experience on liposome-based interaction analyses. A further focus in the group is the in vivo analysis of endosomes and vacuoles, and their association with protein complexes to clarify protein dynamics and function within the endosomal system.
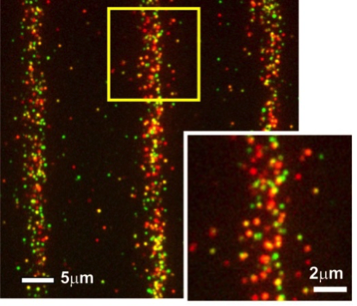
Single cell pull-down assay of individual cytosolic STAT1 (red)/STAT2 (green) complexes on a micropatterned surface (Wedeking, et al., Nano Lett. 2015)
You lab, Biology
Surface biofunctionalization for quantification and manipulation of cell signaling
The You group biofunctionalizes nanoparticles and planar substrates for the functional immobilization and lateral organization of proteins on surfaces. Monofunctionalization of quantum dots was successfully implemented for probing molecular and cellular determinants of cytokine signaling at single molecule level. Micropatterned biofunctionalized surfaces were developed for quantifying dynamics and stoichiometry protein-protein interactions involved in downstream signal propagation of cytokine receptor signaling. Currently, these tools are being expanded to spatiotemporally control the organization of signaling complexes in the plasma membrane of live cells.

